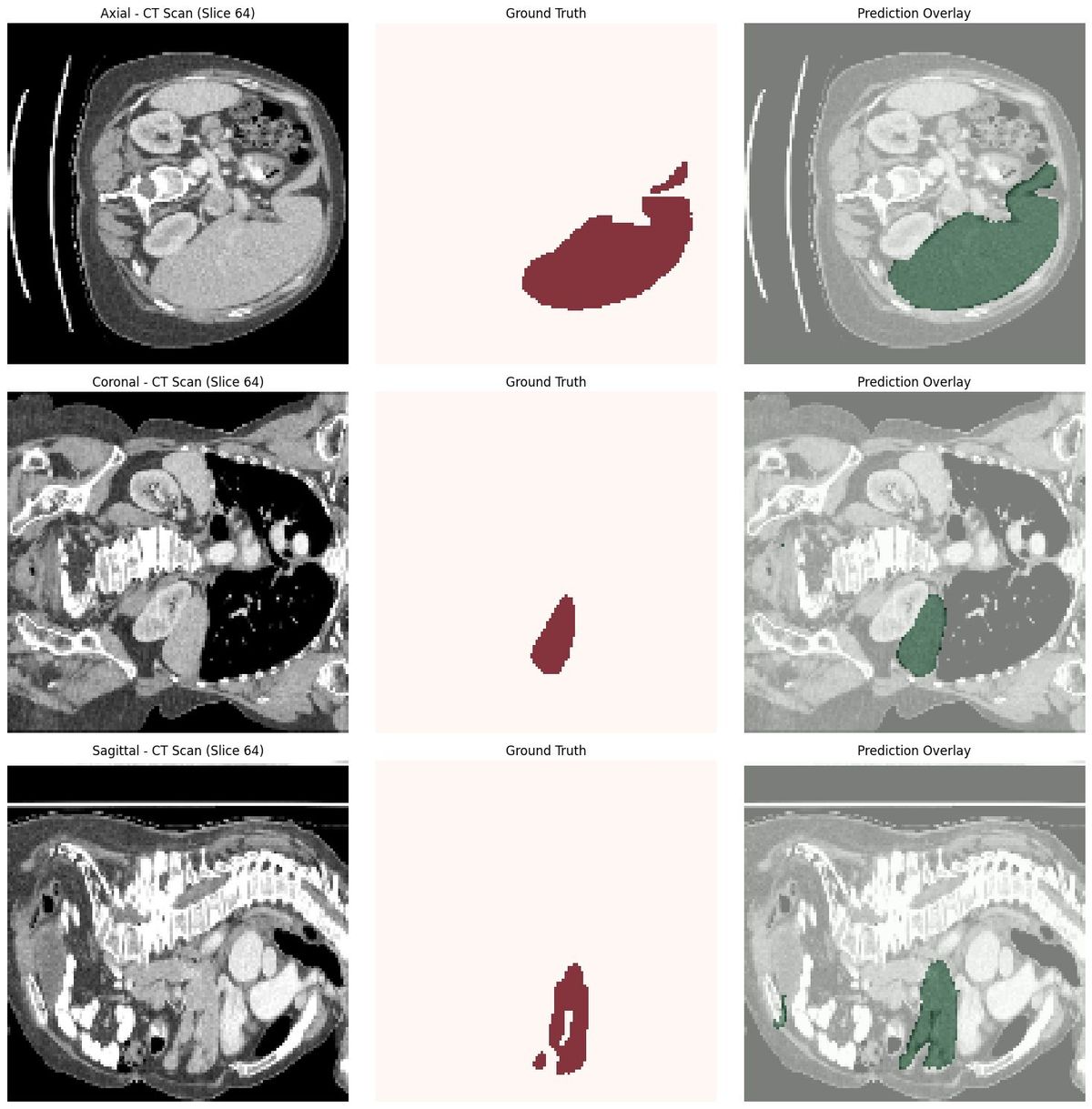
Liver CT Segmentation- Using Monai
This was my first experience working with a large-scale 3D medical imaging dataset. The MSD Task03 Liver dataset is over 25GB, and training a full 3D model required designing a strong backend pipeline for data preprocessing, batching, and sliding-window inference.
Training the model for ~160 epochs on my local RTX 3060 (12GB), and the system achieved a 95.6% Dice score.
SwinUNETR captures long-range anatomical dependencies better than CNNs.
What the system does:
1. Reads and processes full 3D abdominal CT volumes
2. Standardizes orientation (RAS), voxel spacing, intensity ranges, and crops foreground
3. Uses a 3D Vision Transformer (SwinUNETR) for segmentation
4. Handles inference using MONAI’s sliding-window engine and produces accurate 3D liver masks that can be downloaded as NIfTI files
What I built:
1. A fully modular preprocessing pipeline (orientation, cropping, resizing)
2. An optimized training loop with Dice loss, mixed precision, checkpointing, and metric tracking
3. A complete evaluation script for volume-wise Dice scoring
4. A trained SwinUNETR model that generalizes well on held-out test CT volumes